Lifetime-Fitting Using the FLIM Analysis (updated for SymPhoTime V 2.5 and above)
Introduction
This tutorial shows step-by-step, how the FLIM script of SymPhoTime 64 (V2.5) can be used to perform a pixel-by-pixel lifetime fit of a FLIM image and how to extract and read the results. In detail, the FLIM script is started using an image of mouse kidney tissue in the “Tutorials” workspace,https://figshare.com/s/4957fcfa684daef86c23, a suited fitting model is chosen and a pixel-by-pixel fit is performed.
Step-by-step Tutorial
Select a file and start the analysis
- Start SymPhoTime 64 software version (version 2.5 is used for this tutorial, there may be slight changes in other SymPhoTime versions).
- Open the “Tutorials” workspace via File → Open Workspace from the main menu.
Response:
The files of the sample workspace are displayed in the workspace panel on the left side of the main window.
- Highlight the file
Kidney_Cells_FLIM.ptuby a single mouse click. - Select the “Analysis” tab and in there, open the “Imaging” drop-down menu.
Note:
The drop-down menu can be opened and closed by clicking on the grey button on the left side (small set of arrows) of the header of the drop down menu or by clicking onto the “Analysis” header:
- Start the FLIM analysis by clicking on “Start”.
Response:
The file Kidney_Cells_FLIM.ptu is opened with the FLIM analysis. Thereby, a new Window opens:
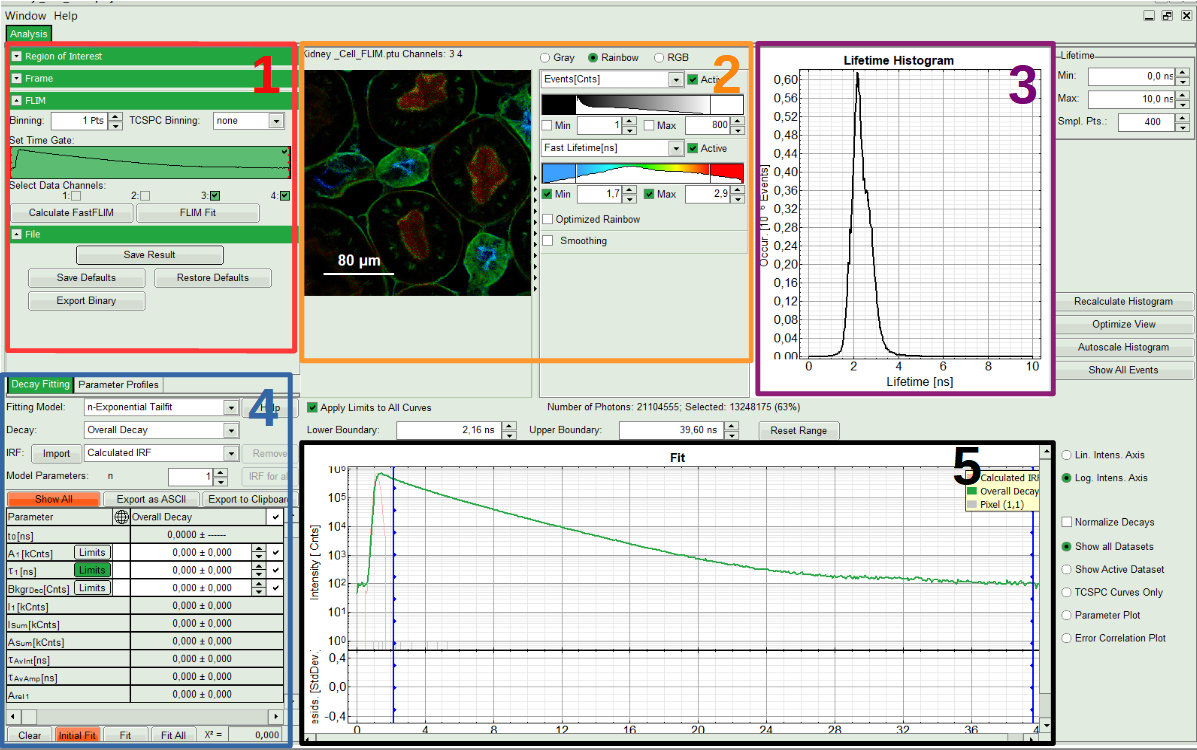
Note:
The window contains five different sections:
1: Setting panel
Imaging analysis options can be defined in this section.
2: Fast FLIM image
The FLIM preview image displayed in a false color scale. The brightness encodes the intensity, while the color encodes the average “Fast FLIM” lifetime, i.e. the mean arrival times of the photons after the laser pulse. When not defined otherwise, intensity and color scale stretch from minimum to maximum. As the mean arrival time of the background photons in the areas where no fluorescence is detected, is randomly spread over the TCSPC window, the mean photon arrival time of the dark background is usually very long (with an average up to 1/2 of the TCSPC window), which makes the color scale loaded by default sometimes unsuited for displaying the lifetime contrast in the actual sample. In this case, the scale can be adapted.
3: Fluorescence Lifetime Histogram
Here the frequency of photon counts corresponding to the individual mean lifetimes is plotted. The settings of the plot can be adapted using the controls on the right of the plot. The histogram is photon weighted.
4: Decay Fitting Parameters
Here the lifetime fitting model can be selected. Fitting is then applied to the lifetime graph(s) on the lower center (TCSPC histogram).
5: TCSPC Decay window
By default, this set shows the TCSPC histogram of all photons in the image in green (= dataset 0) and the TCSPC histogram of a single pixel in grey (= dataset 1). In red an estimation of the instrument response function (=IRF) is shown. The IRF reconstruction is deducted from the rising edge of the TCSPC histogram. Check the SymPhoTime online help for more details.
- To enhance the lifetime contrast, adapt the color scale by typing the limit values for the “Fast Lifetime [ns]” and “Events[cnts]”. If you wish, you can set the “Lifetime Histogram” range on the right hand.
- Tick the checkbox of suited detection “Data Channels” (i.e. detector), in this case 4, and press “Calculate FastFLIM”.
Response:
FLIM Preview image will get updated. Optimize contrast by adapting “Min” and “Max” values.
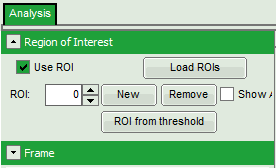
Select ROIs
- Open the “Region of Interest” drop-down menu, tick “Use ROI” checkbox and click “ROI from threshold”.
Response:
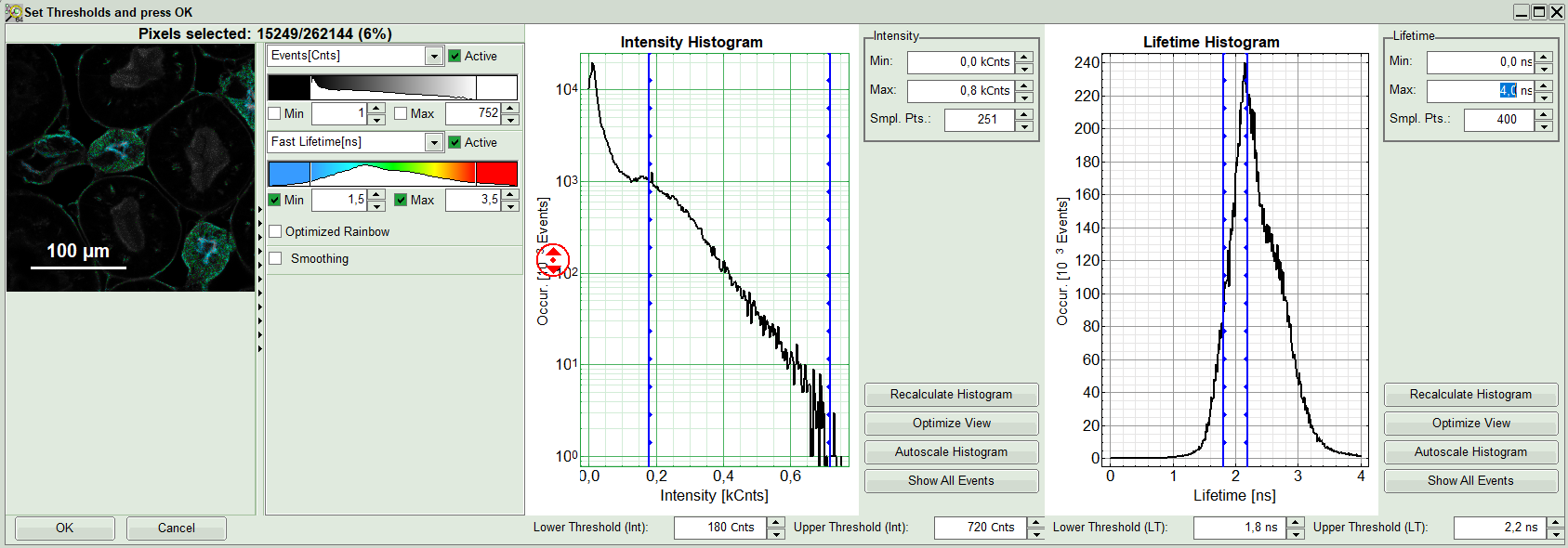
A large window with three sections will appear, where you can set a threshold for an image analysis visually.
Note:
Left to right: Preview FLIM Image, Intensity Histogram and Lifetime Histogram. For any ROI you can set each of these parameters individually. There are two ways to define the new threshold. You can use the edit-box at the lower side to type the threshold and press enter to check the result or use cursor keys or mouse wheel to increase/decrease the value. Another option is using the blue vertical bars on the sides of the intensity or lifetime histograms; click and drag the bars to set the threshold.
- First adapt the contrast by setting the Number of Events [Cnts] which you minimally want to have in a pixel in order to perform a fit. If there are too few photons in a pixel, a decent fit result cannot be expected. It is also possible to additionally set a maximum intensity threshold, if you want to exclude very bright pixels. In this example however, we just set a lower intensity threshold.
- Additionally, a lifetime threshold can be defined. Let's assume that in this example of the kidney cells we're only interested in the blue and green regions of the image (nuclei and elements of glomeruli). In that case we can exclude the cells with a very long lifetime shown in red by moving the bar. A practical application would be to e.g. just analyze transfected, but not autofluorescent cells. Check the image on the left, as you move the bars.
- Optionally, you can exclude unwanted part of the image, if there is any; for instance here using “Free ROI” tool from the context menu and
<CTRL>button, we remove the structures appeared on the left side of the image.
- When you are content with the selection, press OK to select these pixels of the image as Region of Interest (ROI 0).
Response:
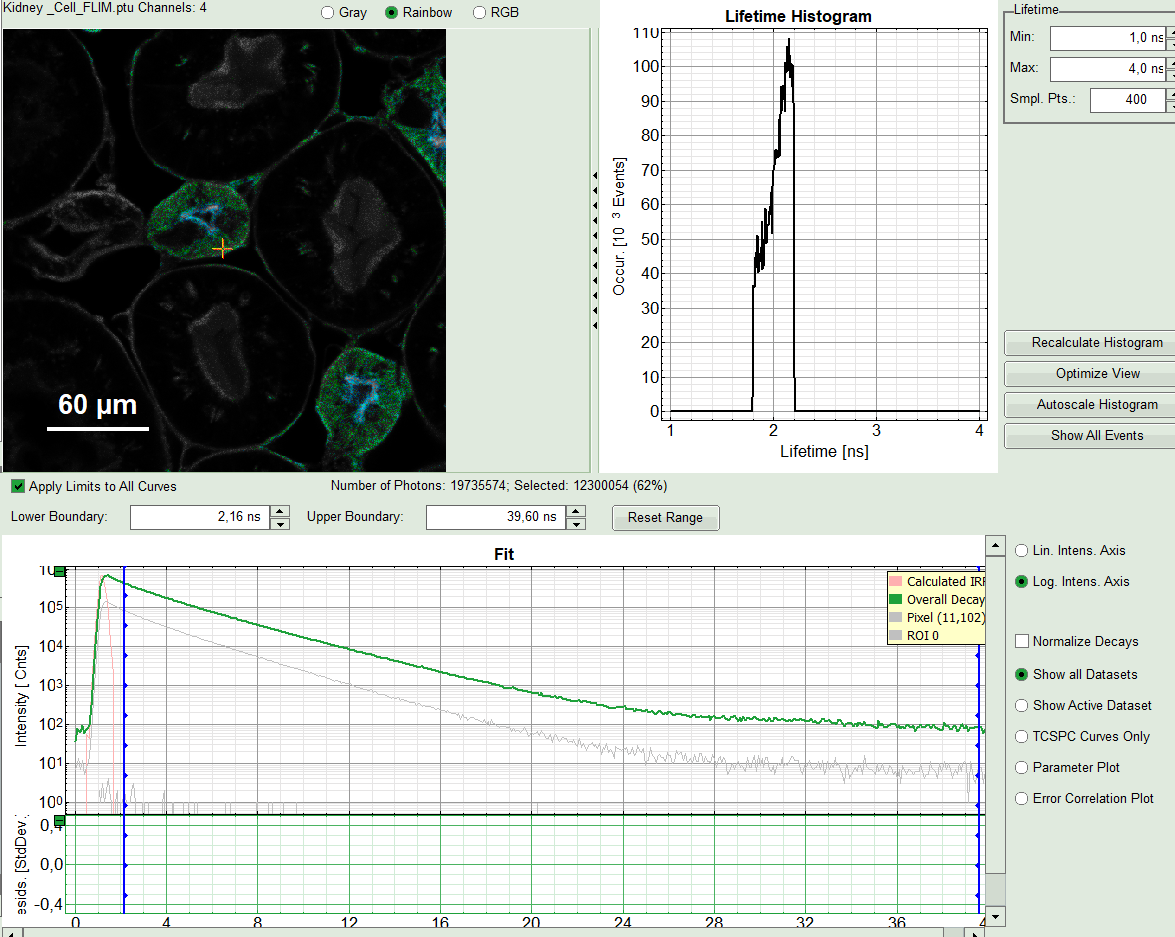
FLIM preview image of ROI 0, as well as its Lifetime Histogram will get updated. A corresponding decays with gray color appear in TCSPC window under “Overall Decay”.
Select the fitting model and determine start parameters
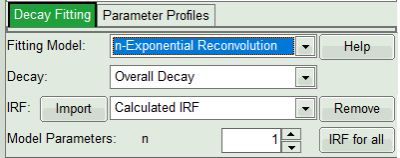
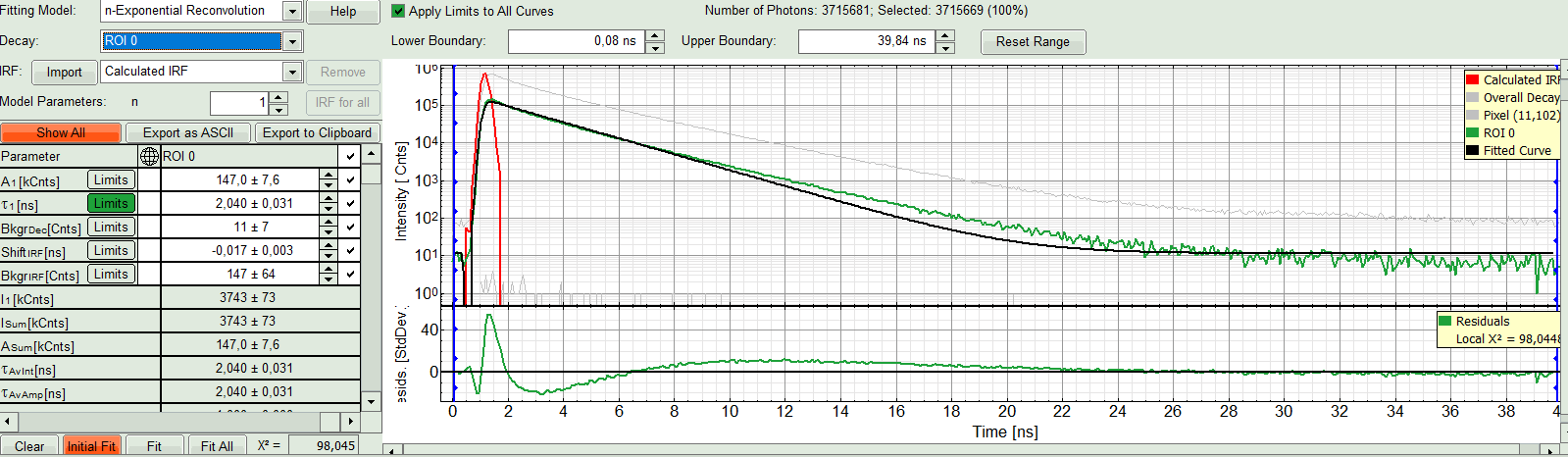
- In the fourth box, “Decay Fitting”, below the image, select the Fitting Model drop-down list select “n-Exponential Reconvolution Fit”.
Response:
In the TCSPC window, the IRF is displayed in bright red, the data fitting limits are moved to the border of the TCSPC window.
The new fitting parameters “Shift IRF” and “Bkgr IRF” (=background IRF) appear in the fitting parameter table.
Note:
The software offers the possibility to fit the data using a n-exponential tailfit, n-exponential reconvolution or rapid reconvolution fit. A tailfit can be used when the fitted lifetimes are significantly longer than the instrument response function. A reconvolution fit is usually preferable, because the complete decay is fitted, while for a tailfit, the start of the fitting range is usually a bit arbitrary. A rapidReconvolution fit is needed, if the count rates, with which the image was acquired are significantly above the Pile Up limit. It is also needed, if the pulse frequency is too high for the fluorescence decay to decrease to the background level before the end of the decay window (often, this is the case for 2-Photon-Excitation data acquired with a TiSa-laser with 80MHz repetition rate). Check out the rapidReconvolution tutorial for this case (https://www.tcspc.com/doku.php/howto:lifetime-fitting_using_the_rapid_reconvolution_algorithm).
For explanations about the fitting model and the used equations, click on the “Help” button next to the selected model. This opens a help window containing the fitting equation and the explanation of the different parameters.
- If you want to use a measured IRF, press on “Import” IRF button and select the corresponding file from the file list (make sure that the IRF is taken from the detector you analyse here only, with the same repetition rate). Click “IRF for All”. In the example here, however, we will work with the estimated IRF automatically generated from the data set. Check the IRF-related tutorials, if you want to work with a measured IRF.
- From “Decay” drop-down list select
ROI 0and click “Initial Fit” (marked in orange).
Response:
A single exponential reconvolution fit is performed. In the TCSPC window, the fit is displayed as a black line. Below, the residuals (= difference between raw data and fit values) are displayed.
- Fit values appear in the fitting table. Check the fitting curve, residuals and χ².
Note:
Usually, a decent fit is characterized by the following criteria:
The fitted curve overlays well with the decay curve. In the residual window, the values spread randomly around 0.
The χ²-value approaches 1.
The calculated fitting values are reasonable.
The fitting model with least parameters is preferred.
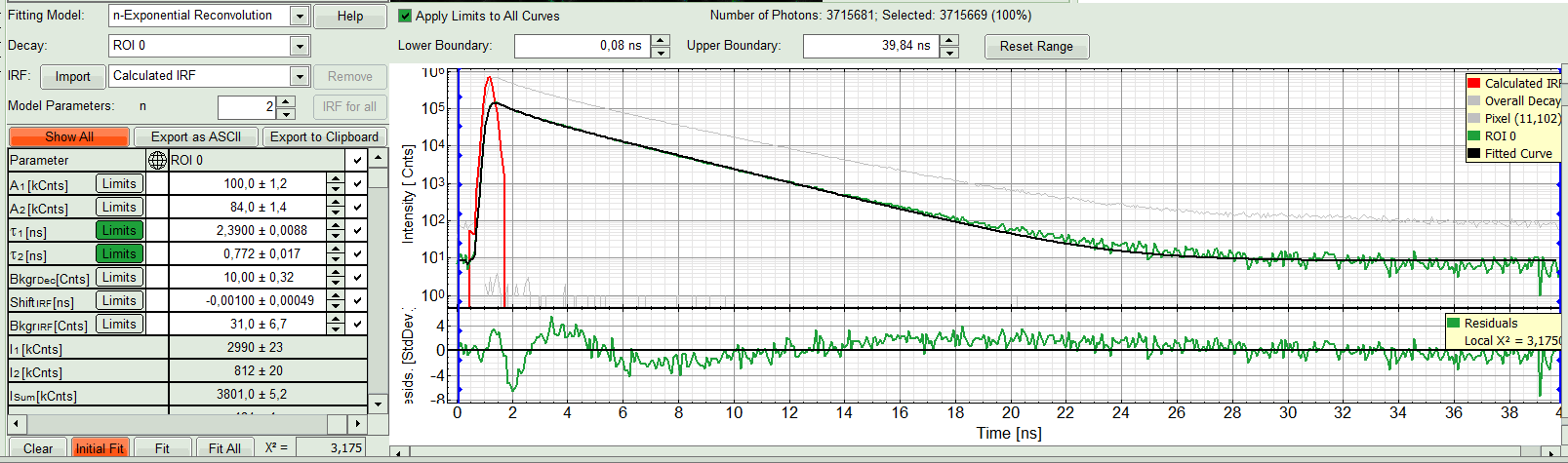
- In this example it is obvious that the applied single exponential reconvolution fit is not suited to fit the decay curve. Increase the “Model Parameters” to n = 2. Again, click on “Initial fit”.
Response:
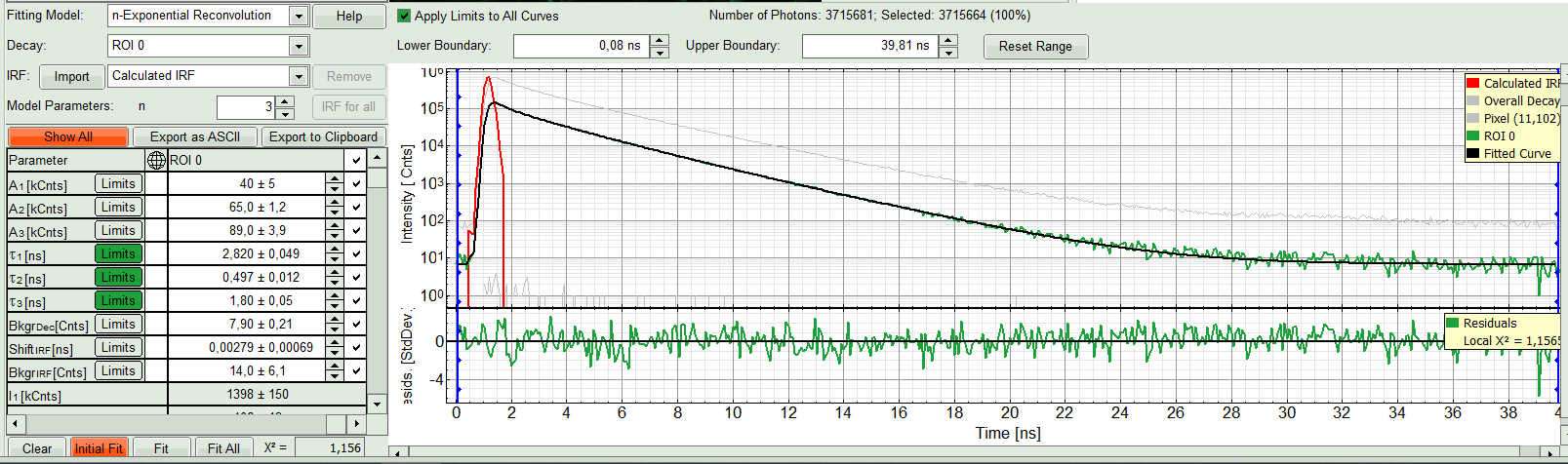
The fit quality increases. But if it still doesn't look sufficient, increase number of “Model Parameters” to three.
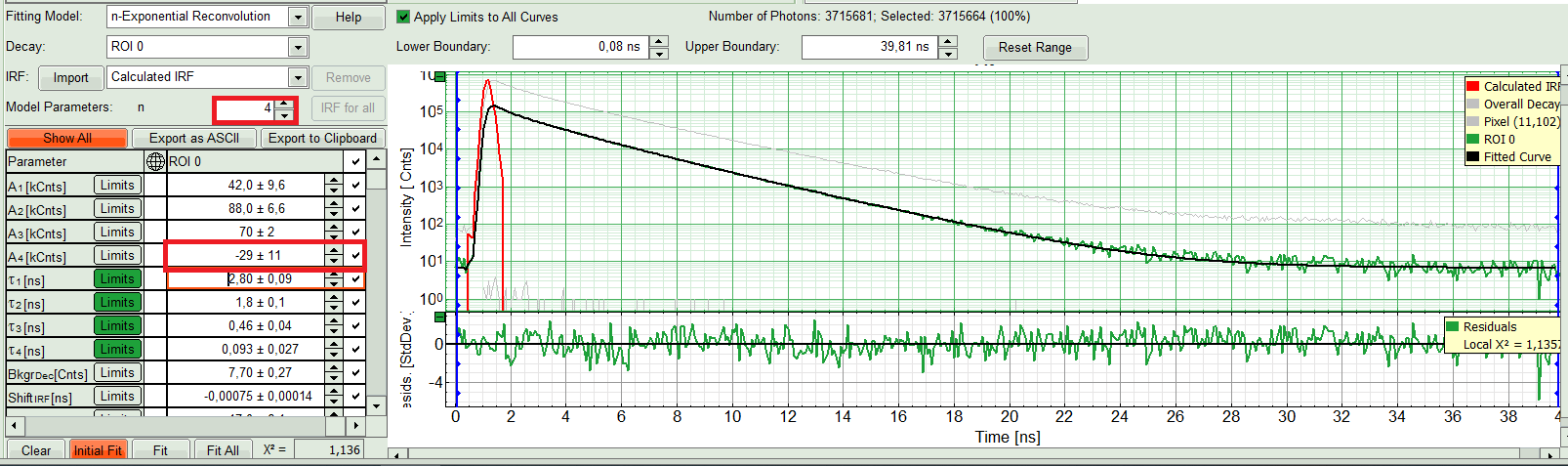
- Three components exponential decay seems to be suitable for this ROI. However we can try four exponential decay and see the result:
Response:
The fourth component appears with a negative amplitude, which is physically not possible. Also, the fit quality has apparently not increased. That means a three component exponential fits the best in this case. So we get back to n=3.
Note:
For pixel-by-pixel fits, we are interested about how the lifetime parameters are distributed in the image. ROI fitting for this analysis is the first step to determine the number of fitting parameters here to be used in the pixel-by-pixel fits.
Perform Fit and save results
Note:
The aim of this procedure is to minimize the number of variable parameters, as every variable parameter also adds noise to the fit. It depends on the sample, to which extend this can be done. In order to increase the fitting quality however, we assume that
- The excitation pulse does not shift within the image acquisition time within the TCSPC window, therefore we fix the parameters “Shift IRF” and “Bkgr IRF”. This is practically always valid. In case of an IRF estimated from the decay, these parameters can also be set to 0, as the IRF was derived from the decay.
- As we can see from the pixel decay in grey, the background before and after the decay is practically 0.Therefore, we can also fix this parameter “Bkgr Dec”to 0. This is also very often valid. By default, the pixel in the upper edge is selected (coordinates 0/0). Click on any other pixel to show the pixel decay on this point.
- In this example, we fix the three fitted lifetimes. If the sample contains only three fluorophores, this is also a valid approach, as long as the lifetime of the components does not depend on the local environment within the sample. This is only possible, if no changes of the individual lifetime components are expected.
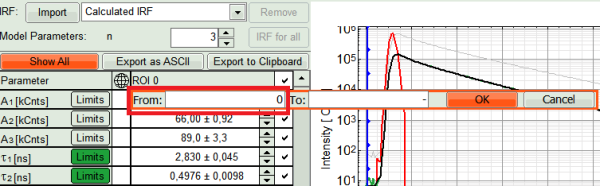
- We also can set constraints on the Amplitudes, to prevent them to run into negative values on the pixel by pixel fit, as the photon statistics for a single pixel are much worse than for the overall ROI. Click on “Limits” button next to A1 in fitting table. Set the lower limit to zero and press OK.
- Set the same limit for A2 and A3.
- Uncheck the boxes for the individual lifetime components, Bkgr(Decay), Bkgr(IRF) and Shift(IRF) in order to fix these values, IRF and Background. Then in the pixel-by-pixel fit, these values are kept constant. Do NOT click “Initial Fit” or “Fit” at this stage!
- Click “FLIM Fit” in the upper panel.
Response:
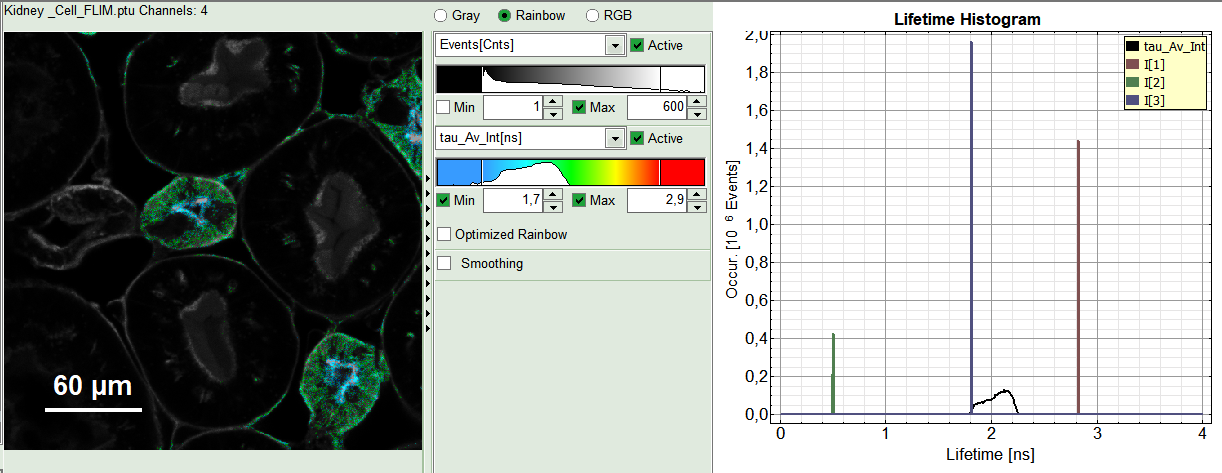
A 3-exponential reconvolution fit is performed on each pixel of “ROI 1”. The result is:
- An updated FLIM image. The colors can slightly change, as now the calculated intensity weighted average lifetime “tau_Av_Int” is displayed, in contrast to the “Fast Lifetime”, which is just the mean arrival time and contains e.g. background photons.
- An updated lifetime frequency histogram on the upper right. The individual lifetime components are displayed as vertical bars, as they are not varied in the pixel-by-pixel fit.
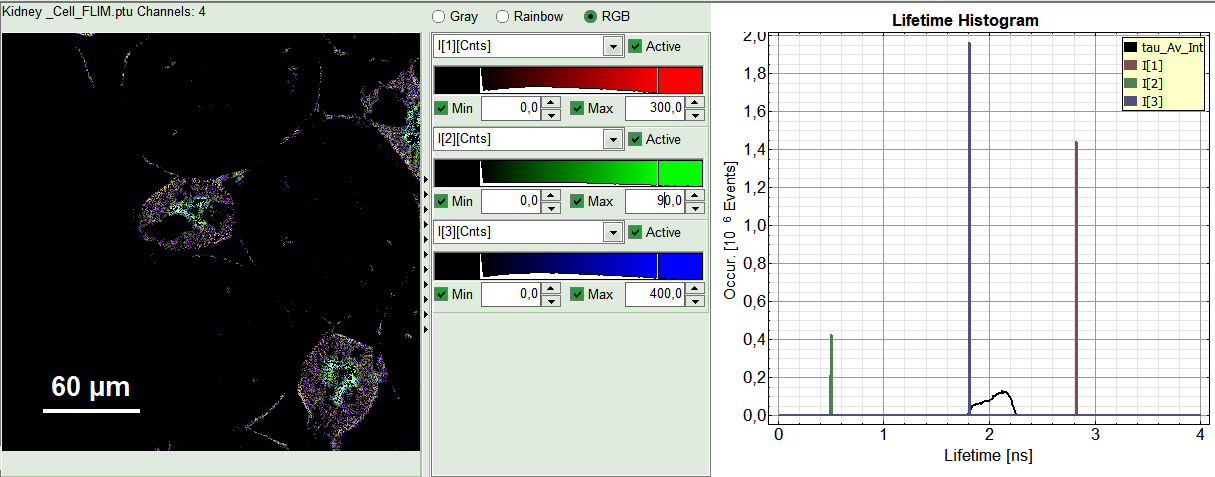
- The three lifetime components can be displayed in a false color scale. Therefore switch the display to “RGB” (above the intensity and color scale) and adapt the intensities of each color as preferred.
Note:
I[1], I[2], I[3] correspond to τ[1], τ[2], τ[3], respectively in the TCSPC fitting panel. It is also possible to plot the amplitudes instead of the intensities or to switch off individual components. You can adapt the intensities of each color as preferred by modifying the displayed false color scale.
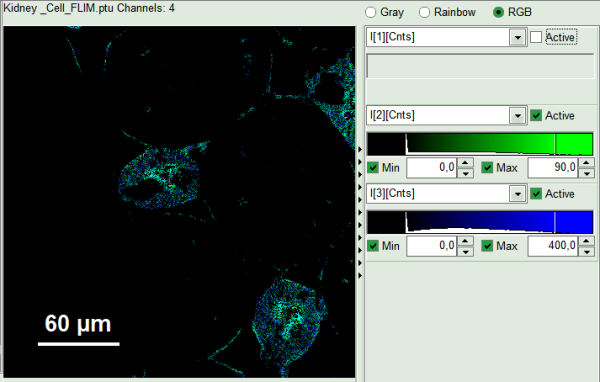
- In RGB mode you can select suited color to your favorite parameter. This helps to distinguish the lifetime of each specious or compartment of the cell. For instance lifetime in nuclei is shown here:
- When the desired display is set, click “Save Result”.
Response:
A result file “FLIM.pqres” is stored under the raw data file Kidney_Cell_FLIM.ptu.
- Later, clicking on the result file reopens the file in the same way as it was stored.