Calculate FCCS Traces with the Grouped FCS Script
Summary
This tutorial shows step-by-step, how to calculate a spectral cross-correlation curve and the corresponding autocorrelation curves of a dye-labeled DNA-oligonucleotide using the Grouped FCS script.
Background Information
Fluorescence Cross-Correlation Spectroscopy (FCCS) is a method to determine molecular interactions between two molecules labeled with different dyes. Usually, the dyes attached to the two molecules of interest differ in their absorption and emission properties. They can also vary in their fluorescence lifetime, for more information please check the tutorials for Fluorescence Lifetime Correlation Spectroscopy (FLCS).
To quantify the cross-correlation signal and connect it to a binding fraction, a calibration of the effective confocal volumes for each dye and the overlapping confocal volume must be performed.
The calibration can be done using a molecule labeled with both fluorophores.
FCCS calibration dyes are available e.g. from IBA (http://www.iba-lifesciences.com/details/product/5-0000-604.html).
Step-by-Step Tutorial
Select a file and start the script
- Start SymPhoTime 64 software.
- Open the “Samples” workspace via “File\open Workspace” from the main menu.
Note: The “Samples” workspace is delivered with the SymPhoTime 64 and on the DVD-ROM and contains example data to show the function of the SymPhoTime 64 data analysis. If you haven't installed it on your computer, copy it from the DVD onto a local drive before going through this tutorial.
Response: The files of the sample workspace are displayed in the workspace panel on the left side of the main window.
- Highlight the file
IBA488+547_crosslinked.ptuby a single mouse click.
Note: This file contains a FCS measurement of a FCCS positive control, a DNA oligonucleotide labeled with a green and a red fluorophore. The sample was excited with a pulsed 485 nm laser and a cw 560 nm laser.
- Select the “Analysis” tab and in there, open the drop down menu “FCS”.
Note: The drop-down menus can be opened and closed by clicking on the grey button on the left side of the header of the drop down menu: 
- Start the Grouped FCS script by clicking on “Start”.
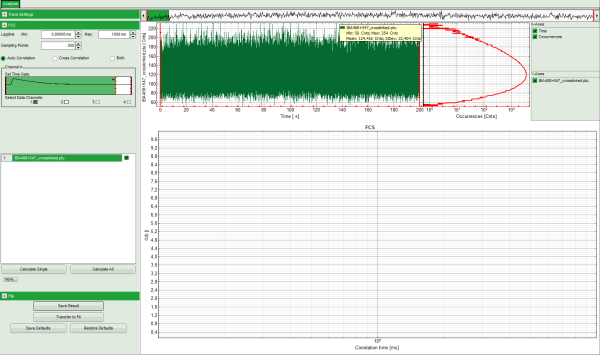
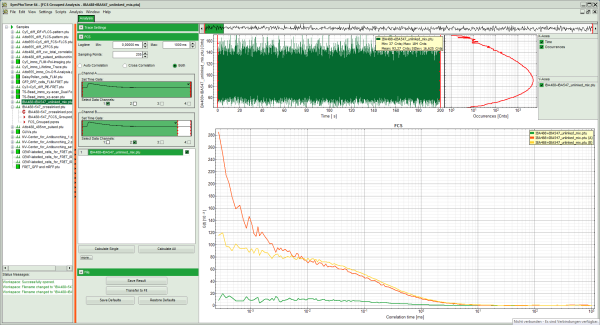
Response: The Grouped FCS script is applied to the file IBA488+547_crosslinked .ptu. Thereby, a new Window opens:
Note: The window contains three different regions:
- Left: Analysis and display options. For explanation of the different parameters, place the mouse cursor over this part of the window and press <F1> to open the corresponding help page. The grouped FCS file can calculate and handle multiple FCS curves, either from different files or several correlations from one file, as used here.
- Upper center/right: Intensity time trace. The display option can be changed using the “Trace Settings” of the analysis options. The large window shows the inset of the complete trace above marked in green. The photon counting histogram on the right displays the frequencies of the different intensity values.
- Lower center/right: FCS-trace(s) window. As first the FCS correlation criteria have to be defined and the curve needs to be calculated, this graph does not contain any trace at this stage.
Calculate the Correlation Curves
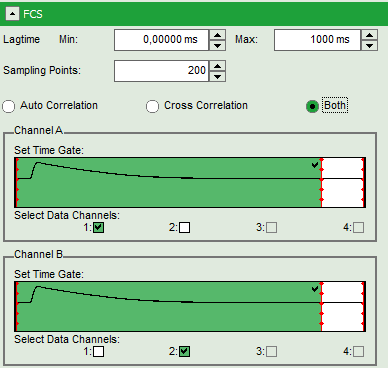
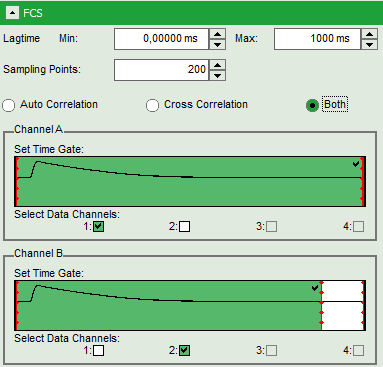
- Set the correlation mode to “Both” to prepare for simultaneous calculation of the autocorrelations and the cross-correlation.
Note: In green, the selected area of the TCSPC window for FCS calculation is marked. It is calculated on the assumption that the sample is excited with pulsed laser light. As for channel 1 (the red detection channel), this is not the case in our sample, we have to select the complete TCSPC window to avoid loosing fluorescence photons.
- For channel A (= detector 1), move the red bar to the right to expand the TCSPC selection over the whole TCSPC window for that channel. The complete TCSPC window should be highlighted in green.
- Press “Calculate Single”.
Response:
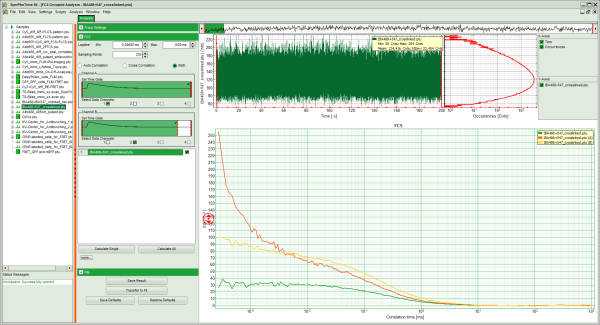
- Three FCS curves are calculated and plotted (the calculation may take some time).
- The first decreasing part of the autocorrelations is due to detector afterpulsing, an unwanted artifact from the detector. As the afterpulsing is not correlated between both detectors, the cross-correlation shows only diffusion behavior.
Note: It can clearly be seen that the cross-correlation is only ~half as high as the autocorrelations. While in autocorrelations, the amplitudes ρ[i] of the autocorrelations correspond to 1/N[i] (= 1 divided by the average number of the molecules in the focus for autocorrelation i=1=Channel A and i=2=Channel B) and are therefore directly linked to the concentrations (C[i]=N[i]/Veff[i]/NA), the relationship is slightly more complex for the cross-correlation: C[1×2]=ρ[1×2]/ρ[1]/ρ[2]/Veff. Ideally, the cross-correlation should match the autocorrelations. Deviations are due to the different effective volumes for both wavelengths and imperfections of the objective, but can also be caused by degraded test samples or too high laser power, which leads to photobleaching of the fluorescent labels.
- Save the result by clicking “Save Results”.
Response: A result file is created.
- If the data should be fitted directly, transfer them to the fitting window by clicking on “Transfer to Fit”. Thereby, the data file is also automatically saved.
- FCS fitting is explained in detail in the tutorial: “Calculate and Fit FCS Traces with the FCS Script”. Look there to find out, how to proceed.
Need for calibration to remove false positive correlation
Note: While the cross-correlation amplitude is decreased from its ideal value due to optical imperfections, spectral crosstalk (usually from the green into the red detector channel) can cause false positive cross-correlation. To measure this effect, a negative calibration probe, i.e. a mixture of two single labeled compounds needs to be measured. To demonstrate this effect, a measurement file of such a sample is provided in the demo workspace.
- Run the same procedure as described before using the file
IBA488+IBA547_unlinked_mix.ptu.
Response:
The correlation curves of this file are calculated. The cross-correlation amplitude of this file is ~20% of the autocorrelations.
Note: In a sample as measured here by a combination of pulsed and cw excitation, it is possible to remove the false positive cross-correlation, which significantly simplifies the binding analysis. This is explained in the tutorial Using the FLCS script for spectral crosstalk removal via FLCCS.